Disease Progression Modeling
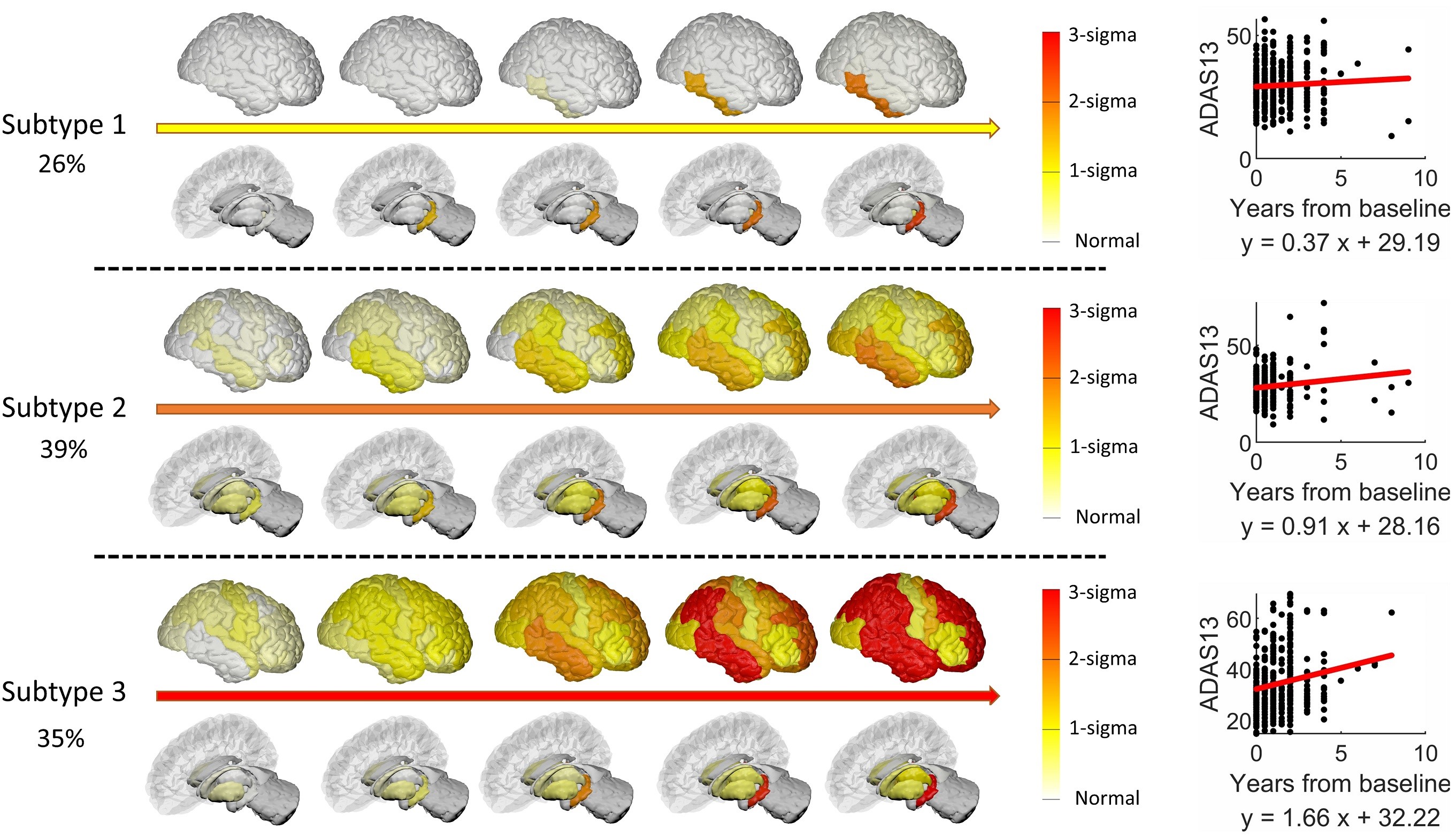
Disease progression models (DPM) aim to recover the trajectory of disease progression using a collection of short time series. A time series corresponds to multiple visits of a patient. At each visit, a patient is examined and various measurements (MRI, PET/SPECT, CSF, clinical scores) about the disease state are acquired. By combining these imaging/CSF/clinical biomarkers from all the patients, the disease progression trajectory can be retrieved. Our research is mainly focused on neurodegenerative diseases, such as Alzheimer's disease (AD), Parkinson's disease (PD) (IPMI 2023, TMI 2021, IPMI 2019).